Excel files
Excel spreadsheets are unique in that they can contain multiple spreadsheets as a workbook.
What an Excel file looks like
Excel file names end with a .xls or .xlsx
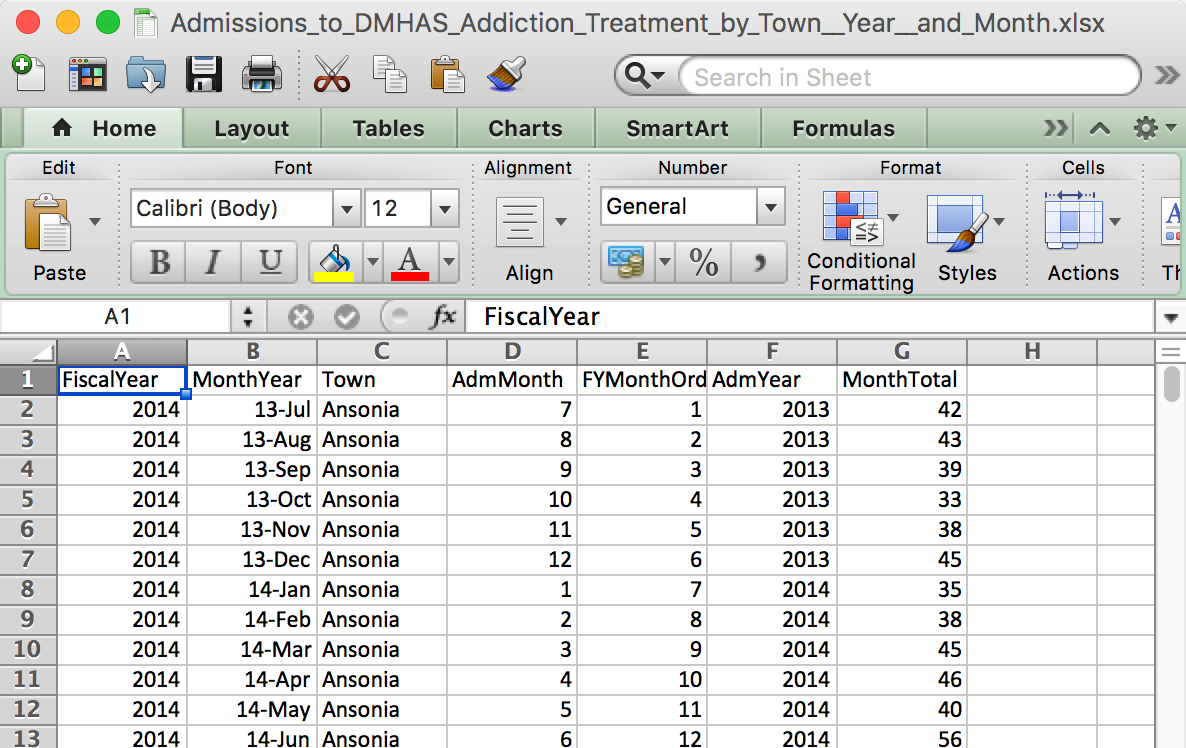
What an Excel file looks like on the inside
Weird, right? Definitely difficult to parse.
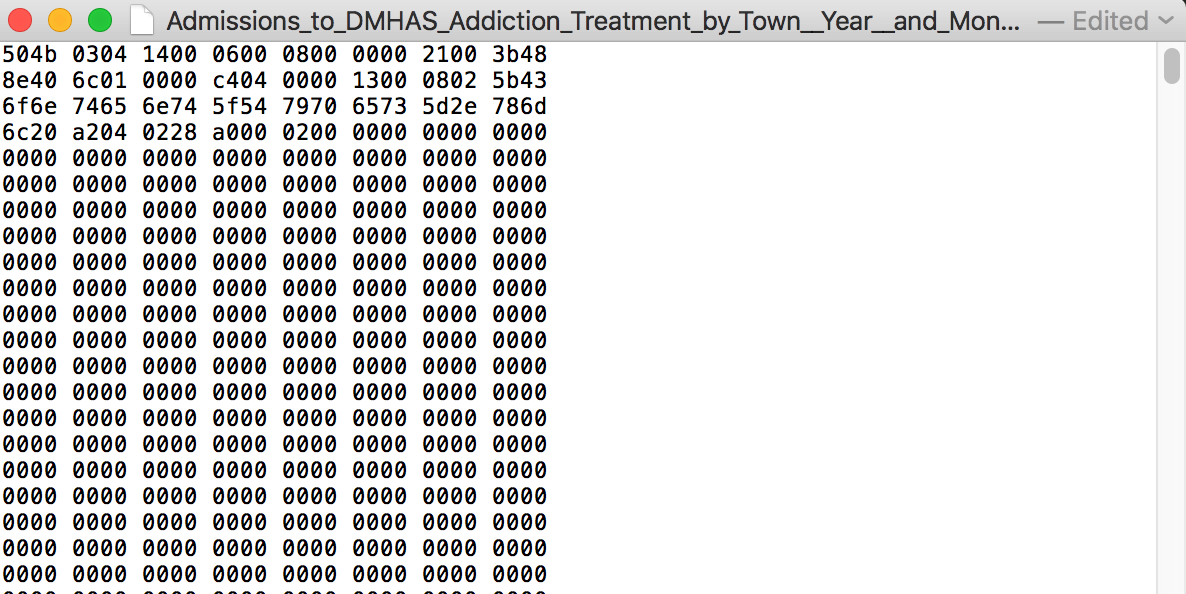
What I do sometimes (often) is save a sheet from Excel as a .csv file.
That’s a short cut. But if you want to be transparent and really deal with the raw data then:
Importing Excel files
- Importing Excel is complicated, readxl package is needed
- There are more other packages that handle Excel files and can build extra sheets, but we won’t be needing them for this instance
Importing Excel files
First, install the readxl package if you have not yet done so.
That will have readxl as part of the group of packages.
## If you don't have readxl installed, uncomment the line below and run it
#install.packages("readxl")
library(readxl)Unlike a csv, you can’t just copy and paste the URL for an Excel sheet.
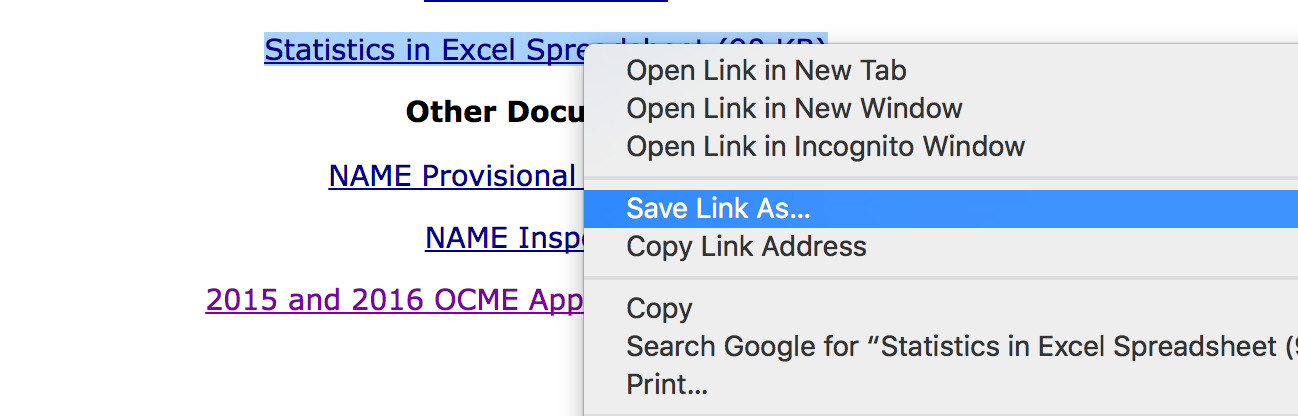
You gotta download the file first.
Right click the link of the Excel data link and click Save File As…
read_excel()
Excel spreadsheets have multiple sheets and it’s best to explore what it looks like in Excel first because read_excel() requires specific sheets to be referred to when importing.
Give it a shot with the first sheet.
df_xl <- read_excel("data/StatisticsSummary.xls", sheet=1)Check it
View(df_xl)
This isn’t right.
The problem with Excel files is that people love to format it in ways that make it look nice in Excel but makes no sense in R.
read_excel() again
But this time we’ll add skip=2 so it skips the first rows when bringing in the data.
df_xl <- read_excel("data/StatisticsSummary.xls", sheet=1, skip=2)Much better
View(df_xl)
Notice that the column names are preserved with spaces and symbols.
# the colnames() function lists the column names of the dataframe
colnames(df_xl)## [1] "Fiscal Year 7/1-6/30" "Accessions"
## [3] "Autopsies" "Exam-inations"
## [5] "Other Cases" "TOTAL"
## [7] "Cremations" "% incl crem"
## [9] "Homicides" "Suicide"
## [11] "Accidents" "Undetermined"
## [13] "ALL" "U 20"
## [15] "U 17" "SIDS"
## [17] "Clinicals"So how would one refer to the data in the columns with spaces
If we did it like normal with the $ to pull the column we’d try
head(df_xl$Other Cases)## Error: <text>:1:18: unexpected symbol
## 1: head(df_xl$Other Cases
## ^See, spaces won’t work. This is how to deal with columns with spaces– add the back tick next to the 1 button on your keyboard.
head(df_xl$`Other Cases`)## [1] 39 17 40 40 40 37It’s some extra finger work that you might be okay with if it was in a limited basis.
However, in anticipation of the work we’re going to be doing, we should go ahead and simplify the column names so there are no characters or spaces. Here’s how
Cleaning (part 1)
We’ll use the make.names() function on the column names. This function makes syntactically valid names out of character vectors (as in in strips out the spaces and replaces them with periods)
colnames(df_xl) <- make.names(colnames(df_xl))Check it
View(df_xl)
colnames(df_xl)## [1] "Fiscal.Year......7.1.6.30" "Accessions"
## [3] "Autopsies" "Exam.inations"
## [5] "Other.Cases" "TOTAL"
## [7] "Cremations" "X..incl.crem"
## [9] "Homicides" "Suicide"
## [11] "Accidents" "Undetermined"
## [13] "ALL" "U.20"
## [15] "U.17" "SIDS"
## [17] "Clinicals"Alright, that’s a bit better.
Still, there’s some oddness in the names but that’s because enters were replaced with periods.
Check out the first column: Fiscal.Year......7.1.6.30
Let’s change that so it’s easier to type later on.
Change the name of a single column
I’ll show you how to do it in Base R and using the dplyr package
Copy Fiscal.Year......7.1.6.30 and paste it into `colnames(dataframe_name)[colnames(dataframe_name) == ‘ColumnNameToBeChanged’] <- ‘NewColumnName’
# Don't run this, I just want to show you the process
colnames(df_xl)[colnames(df_xl) == 'Fiscal.Year......7.1.6.30'] <- 'Year'Here’s how to do it with dplyr: By using the rename() function.
## If you don't have dplyr installed yet, uncomment the line below and run it
# install.packages("dplyr")
library(dplyr)##
## Attaching package: 'dplyr'## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, uniondf_xl <- rename(df_xl, Year=Fiscal.Year......7.1.6.30)It’s slightly different– there are less parentheses and brackets and equal signs.
And you don’t need to add quotation marks.
Check it
colnames(df_xl)## [1] "Year" "Accessions" "Autopsies" "Exam.inations"
## [5] "Other.Cases" "TOTAL" "Cremations" "X..incl.crem"
## [9] "Homicides" "Suicide" "Accidents" "Undetermined"
## [13] "ALL" "U.20" "U.17" "SIDS"
## [17] "Clinicals"Fix the other names if you want. I’m going to leave them as is for now.
Is the df_xl sheet clean enough to work with?
Scroll down to the bottom of the data.

Not clean yet. There are a bunch of NAs.
That might give us some issues later on so let’s take care of it now.
Eliminating NAs
Easiest way to get rid of NAs is to subset or filter out the NAs based on one column.
Let’s use the Year column.
There are two ways: subset() or filter()
- Base R
df_xl <- subset(df_xl, !is.na(Year))- dplyr
## If you don't have dplyr installed yet, uncomment the line below and run it
# install.packages("dplyr")
library(dplyr)
df_xl <- filter(df_xl, !is.na(Year))What’s the difference? dplyr is part of the tidyverse suite of packages that we’ll be getting into later on in the course. Go ahead and use that.
Check it

No NAs at the bottom.
It took a few lines of code but the data has been cleaned up enough to analyze or visualize with.
Exporting to Excel
It’s preferable to save data frames as CSVs because it’s more open and doesn’t require a paid program for others to open.
But if you must, there are some decent walkthroughs:
Your turn
Challenge yourself with these exercises so you’ll retain the knowledge of this section.
Instructions on how to run the exercise app are on the intro page to this section.
© Copyright 2018, Andrew Ba Tran